Development of NMR methods for ‘imaging’ DNA and RNA dynamics
Techniques based on NMR spin relaxation in the rotating frame (R1ρ) and chemical exchange saturation transfer (CEST) are providing rare insights into short-lived (μs-ms) low-populated (<5%) transient conformational states in DNA and RNA that play important roles in gene expression and regulation. I developed the nucleic acid sugar 13C R1ρ experiments to resolve and confirm the structures of two transient states in DNA: transient Hoogsteen base pairs and transient Watson-Crick like G-T mismatches. In addition, I also developed an non-invasive NMR CEST approach for measuring the kinetics of nucleic acid hybridization and discovered that epitranscriptomic modification m6A slows the annealing of RNA duplexes, which explains why in mRNA, m6A slows down tRNA selection.
|
 |
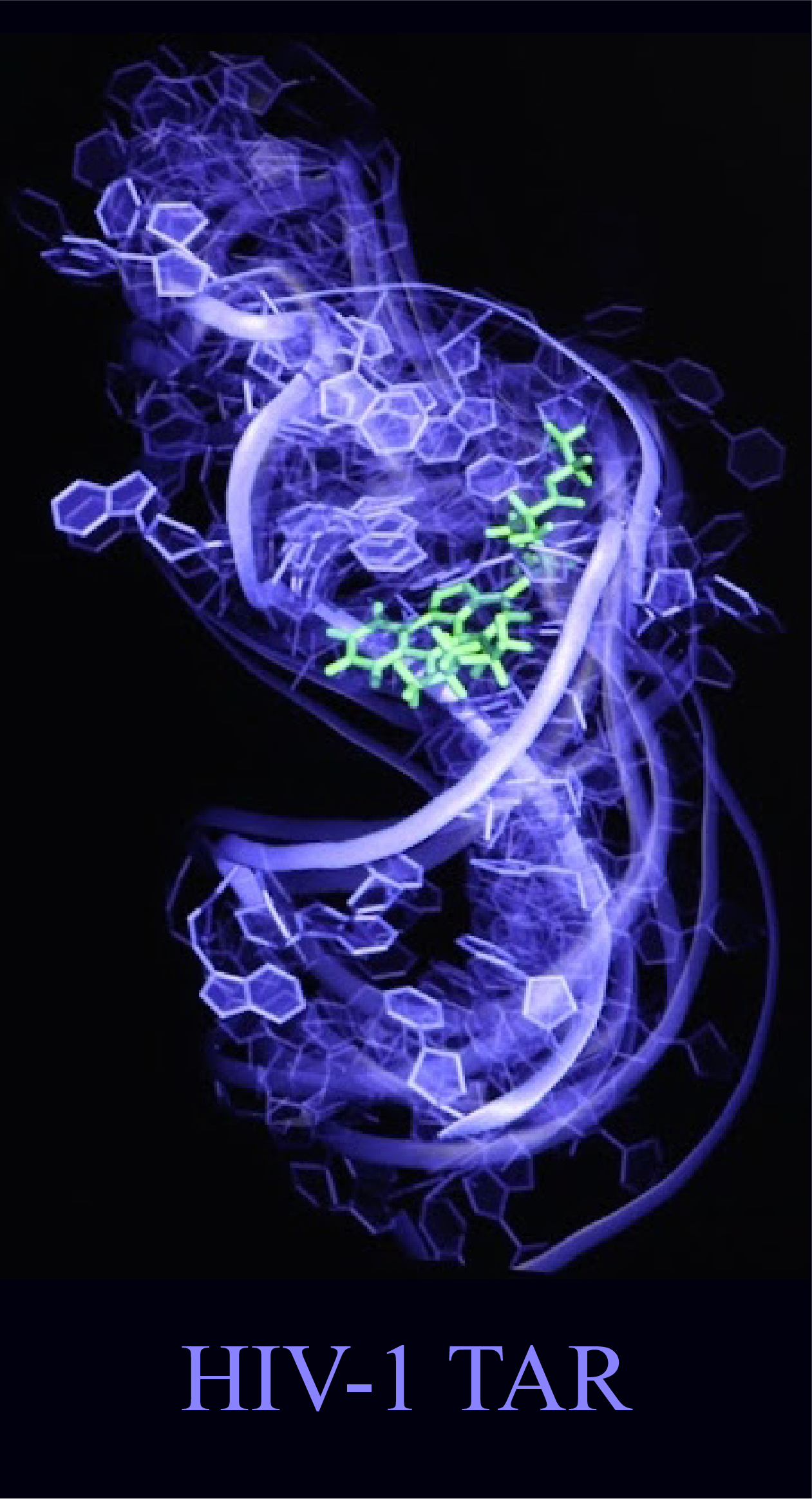
Rapid and accurate determination of RNA dynamic ensembles
Functional and regulatory RNAs do not fold into a single 3D structure but rather form dynamic ensembles of many inter-converting conformations and undergo conformational transitions in multi-step biochemical cycles, ligand binding, and signaling. Solving dynamic ensembles of RNA at atomic resolution is a major challenge in structural biology because it requires information which far exceeds that available from experimental measurements. I addressed the data gap in RNA ensemble determination by combining NMR residual dipolar couplings (RDCs) and a structure prediction tool (Rosetta-FARFAR) that leverages the growing database of RNA structures. The generality of this approach will make determination of atomic-resolution RNA ensembles for RNA-targeted drug discovery routine.
|
 |
Hoogsteen base pairs: Redefining the DNA double helix and genome
In the DNA double helix, Watson-Crick base pairs exist in dynamic equilibrium with non-canonical Hoogsteen base pairs. The Hoogsteen base pairs form through 180º rotation of the purine base and movement of the two partner bases into closer proximity to form Hoogsteen type-hydrogen bonds. I developed a new approach by combining NMR R1ρ and chemical modifications to determine the first high-resolution structure and dynamic ensemble of a DNA double helix containing a Hoogsteen base pair. The structure revealed key structural features of Hoogsteen base pairs: the helix is kinked towards the major groove and the helical diameter is constricted. I also developed a novel high-throughput method combining melting experiments and mutagenesis to map the sequence dependent energetics of Hoogsteen base pairs. Now I am working on re-examining the modeling of Hoogsteen base pairs from protein-DNA X-ray structures. I hope my research will bridge the gap of understanding the roles of Hoogsteen base pairs in genome.
|
 |


