|
|
2021 |
|
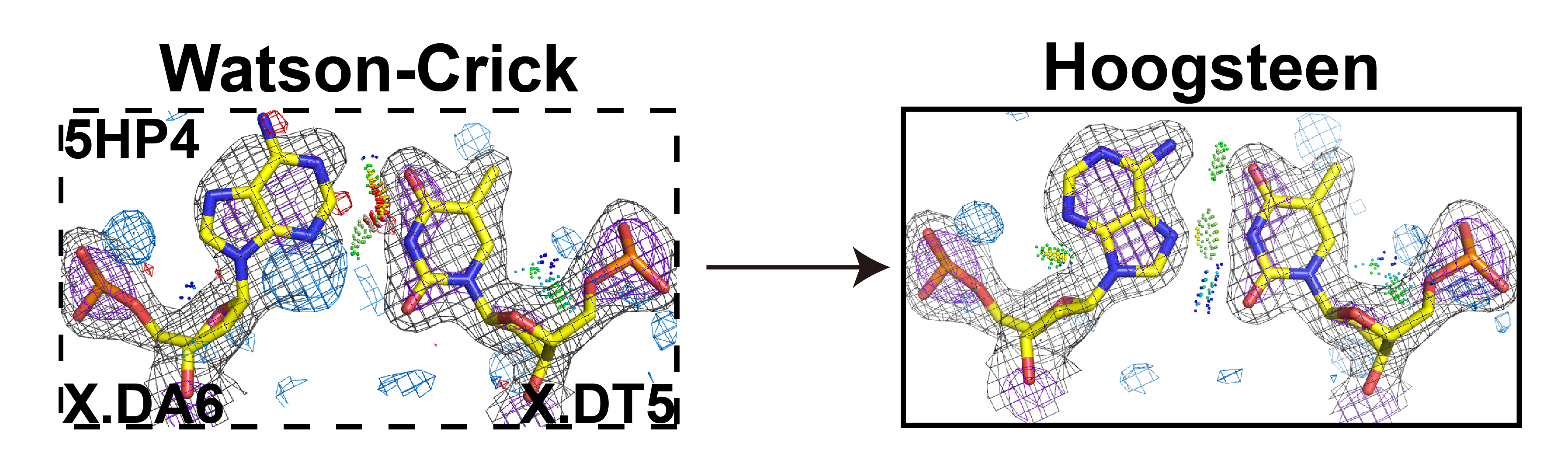
18. Shi H., Kimsey I.J., Liu H.-F., Pham U., Schumacher M.A..*, Al-Hashimi H.M.* (2021) “Revealing A-T and G-C Hoogsteen base pairs in stressed protein-bound duplex DNA” bioRxiv
|
|
|
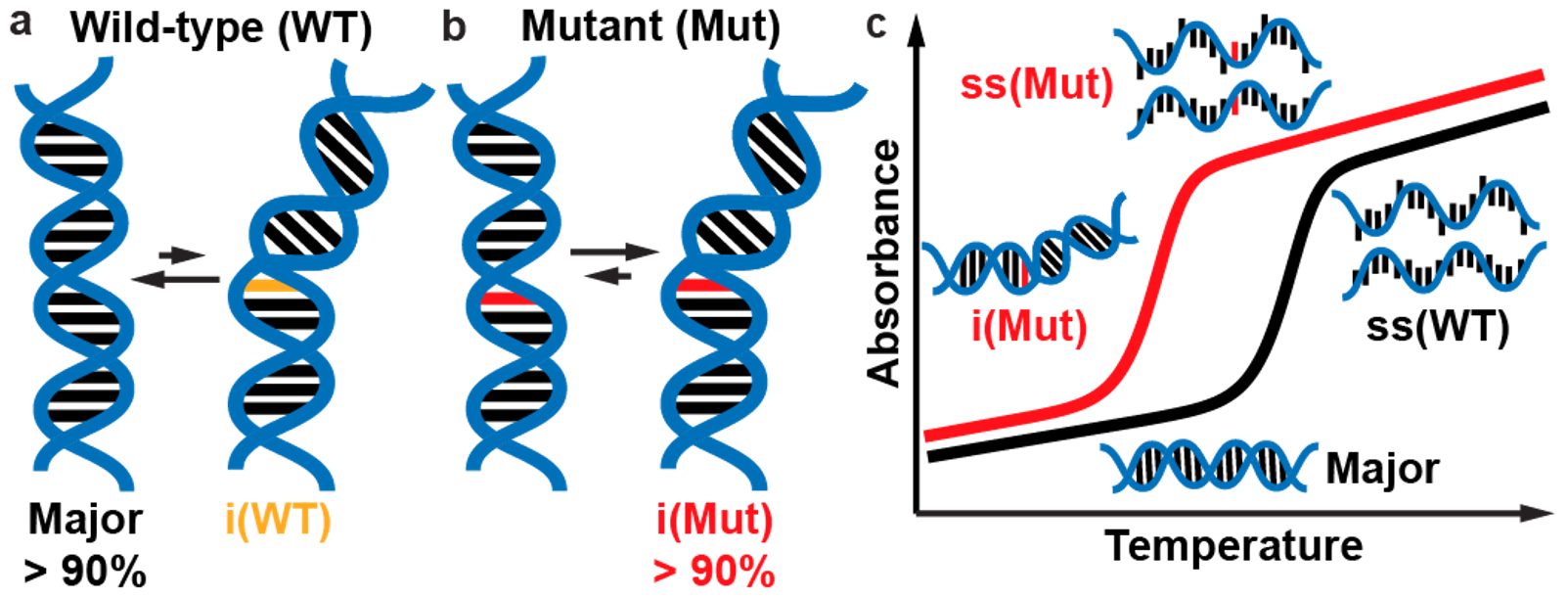
17. Rangadurai A.#, Shi H.#, Xu Y.#, Liu B., Abou Assi H., Zhou H., Kimsey I.J., Al-Hashimi H.M.* (2021) “delta-Melt: Nucleic acid conformational penalties from melting experiments” bioRxiv
|
|
|
16. Liu B., Shi H., Rangadurai A., Nussbaumer F., Chu C-C., Erharter K.A., Case D.A., Kreutz C., Al-Hashimi H.M.* (2021) “A quantitative model predicts how m6A reshapes the kinetic landscape of nucleic acid hybridization and conformational transitions” bioRxiv
|
|
|
15. Liu B., Shi H., Al-Hashimi H.M.* (2021) “Developments in solution-state NMR yield broader and deeper views of the dynamic ensembles of nucleic acids” Current Opinion in Structural Biology 70: 16-25
|
|
|
14. Kelly M.L., Chu C-C., Shi H., Ganser L.R., Bogerd H.P., Huynh K., Hou Y., Cullen B.R.*, Al-Hashimi H.M.* (2021) “Understanding the characteristics of nonspecific binding of drug-like compounds to canonical stem-loop RNAs and their implications for functional cellular assays” RNA 27(1): 12-26
|
2020
|
|
13. Abou Assi H., Rangadurai A., Shi H., Liu B., Clay M.C., Erharter K.A., Kreutz C., Holley, C.L.*, Al-Hashimi H.M.* (2020) “2′-O-methylation can increase the abundance and lifetime of alternative RNA conformational states” Nucleic Acids Research 48(21):12365-12379
|
|
|
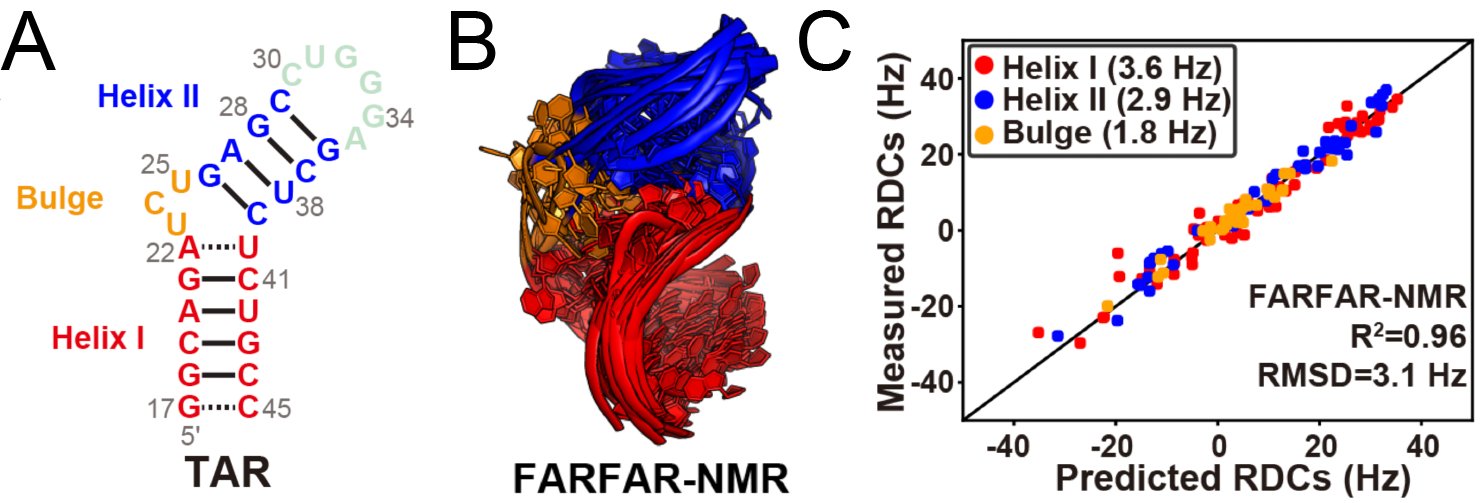
12. Shi H., Rangadurai A., Abou Assi H., Roy R., Case D.A.*, Yesselman J.D.*, Herschlag D.*, Al-Hashimi H.M.* (2020) “Rapid and accurate determination of atomistic RNA dynamic ensemble models using NMR and structure prediction” Nature Communications 11: 5531
|
|
|
11. Xu Y.#, Manghrani A.#, Liu B., Shi H., Pham U., Liu A., Al-Hashimi H.M.* (2020) “Hoogsteen base pairs increase the susceptibility of double-stranded DNA to cytotoxic damage” Journal of Biological Chemistry 295(47): 15933-15947
|
|
|
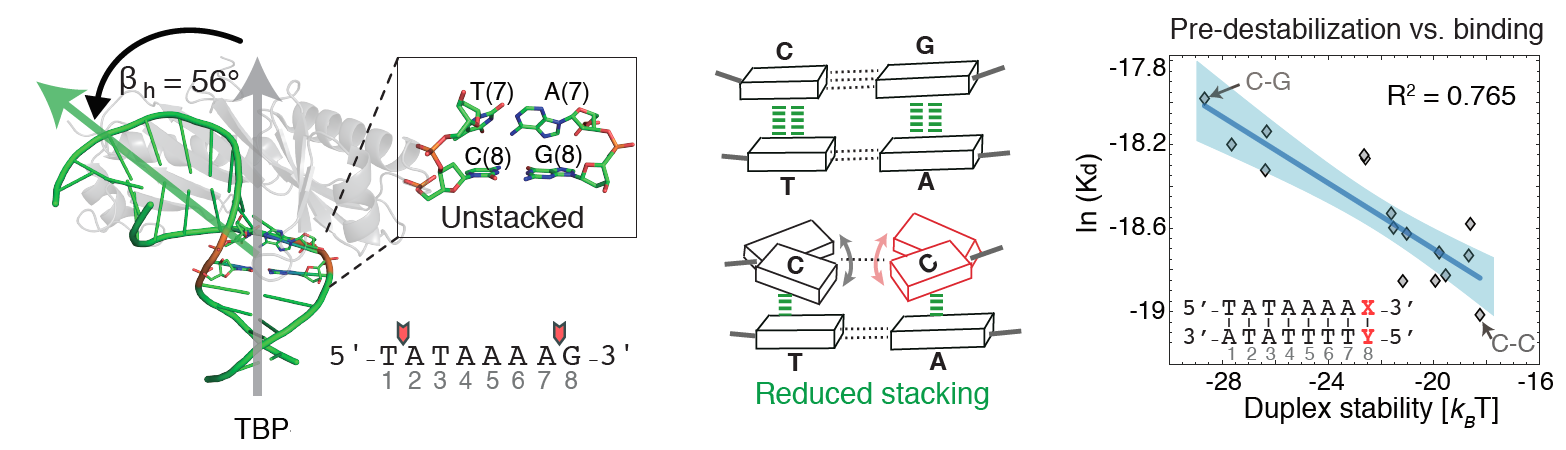
10. Afek A., Shi H., Rangadurai A., Sahay H., Senitzki A., Xhani S., Fang M., Salinas R., Mielko Z., Pufall M.A., Poon G.M.K., Haran T.E., Schumacher M.A., Al-Hashimi H.M.*, Gordan R.* (2020) “DNA mismatches reveal conformational penalties in protein-DNA recognition” Nature 587(7833): 291-296
|
|
|
9. Rangadurai A., Szymanski E.S., Kimsey, I.J., Shi H., Al-Hashimi H.M.* (2020) “Probing conformational transitions towards mutagenic Watson-Crick-like G•T mismatches using off-resonance sugar carbon R1ρ relaxation dispersion” Journal of Biomolecular NMR 74(8-9): 457-471
|
|
|
8. Rangadurai A., Shi H., Al-Hashimi H.M.* (2020) “Extending the Sensitivity of CEST to Micro-to-Millisecond Dynamics in Nucleic Acids Using High Power Radio-Frequency Fields” Angewandte Chemie International Edition 59(28): 11262-11266
|
2019 |
|
7. Shi H.#, Liu B.#, Nussbaumer F., Rangadurai A., Kreutz C.*, Al-Hashimi H.M.* (2019) “NMR chemical exchange measurements reveal that N6-methyladenosine slows RNA annealing” Journal of American Chemical Society 141(51): 19988-19993
|
|
|
6. Rangadurai A., Kremser J., Shi H., Kreutz C.*, Al-Hashimi H.M.* (2019) “Direct evidence for (G) O6··· H2-N4 (C)+ hydrogen bonding in transient G (syn)-C+ and G (syn)-m5C+ Hoogsteen base pairs in duplex DNA from cytosine amino nitrogen off-resonance R1ρ relaxation dispersion measurements” Journal of Magnetic Resonance 308: 106589
|
|
|
5. Rangadurai A., Szymanski E.S., Kimsey I.J., Shi H., Al-Hashimi H.M.* (2019) “Characterizing chemical exchange using off-resonance R1ρ relaxation dispersion: Application to nucleic acids” Progress in Nuclear Magnetic Resonance Spectroscopy 112-113: 55-102
|
2018 |
|
4. Rangadurai A., Zhou H., Merriman D.K., Meiser N., Liu B., Shi H., Szymanski E.S., Al-Hashimi H.M.* (2018) “Why are Hoogsteen base pairs energetically disfavored in A-RNA compared to B-DNA?” Nucleic Acids Research 46(20): 11099-11114
|
|
|
3. Merriman D.K., Yuan J., Shi H., Herschlag D., Al-Hashimi H.M.* (2018) “Increasing the length of poly-pyrimidine bulges broadens RNA conformational ensembles with minimal impact on stacking energies” RNA 24(10): 1363-1376
|
|
|
2. Shi H., Clay M.C., Rangadurai A., Sathyamoorthy B., Case D.A.*, Al-Hashimi H.M.* (2018) “Atomic Structures of Excited State A-T Hoogsteen Base Pairs in Duplex DNA by Combining NMR Relaxation Dispersion, Mutagenesis, and Chemical Shift Calculations” Journal of Biomolecular NMR 70(4): 229-244
|
2017 |
|
1. Sathyamoorthy B., Shi H., Zhou H., Xue Y., Rangadurai A., Merriman D.K., Al-Hashimi H.M.* (2017) “Insights into Watson-Crick/Hoogsteen Breathing Dynamics and Damage Repair from the Solution Structure and Dynamic Ensemble of DNA Duplexes containing m1A” Nucleic Acids Research 45(9): 5586-5601
|
|
|
* = corresponding author
# = co-first author
|




